SC 2005 LBNL Visualization Group Demos/Projects/Presentations
Table of Contents
SciDAC Visualization: Accelerator Modeling
NERSC related projects
Other Projects
SC05 Talks
SC05 Booth Slideshow
SciDAC Visualization: Accelerator Modeling
- Title:
Visualization of Large Particle Data Sets
- Description:
Simulation studies of beam dynamics using large number of particles produce extremely large six dimensional space data sets . It is desirable to store such enourmous datasets efficiently and also to facilitate the sharing of data between different groups that work of particle-based accelerator simulations.
This work shows a simple HDF5 (Hierarchical Data Format) file schema as well as an API that simplifies reading and writing using the HDF5 library. We also show the work under development to build a set of visualization and analytics tools using this schema.
-
Web page for the self-guided demo.
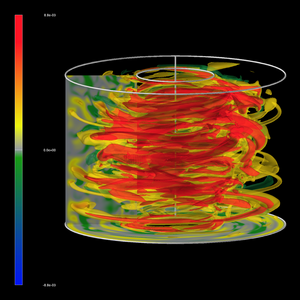
Incite 4: Visualization of Magneto-rotational instability and turbulent angular momentum transport
- Title:
Visualization of Magneto-rotational instability and turbulent angular momentum transport
- Description:
This project studies the forces that help newly born stars and black holes increase in size.
-
Web page for the self-guided demo.
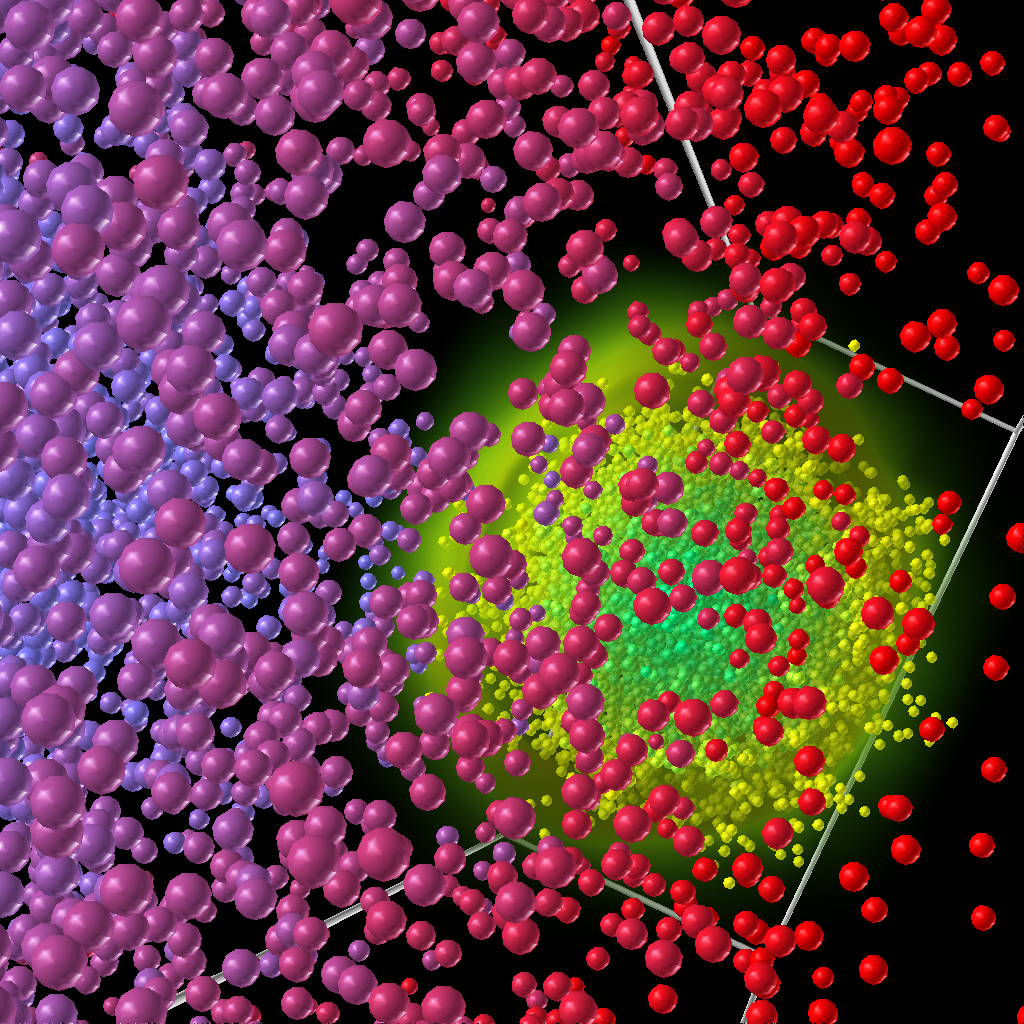
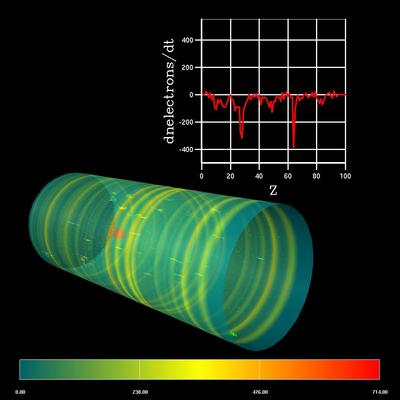
Accelerator Modeling: Electron-cloud Effect
- Title:
Electron-cloud Effect Visualization
- Description:
We continued our collaboration with Andreas Adelmann from PSI to visualize electron cloud simulations. We implemented AVS modules to visualize the charge neutralization along the beam pipe.
-
Web page for the self-guided demo.
Structural Biology: Building Models of Cells
|
- Title:
Building Model of cells
- Description:
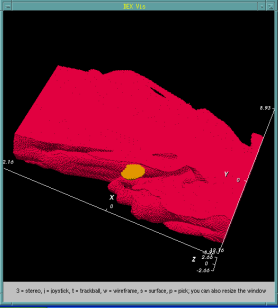
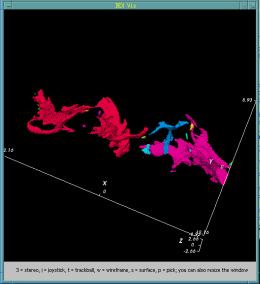
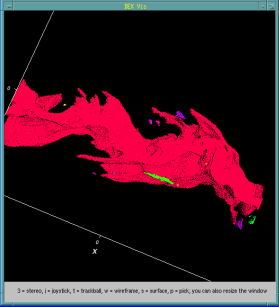
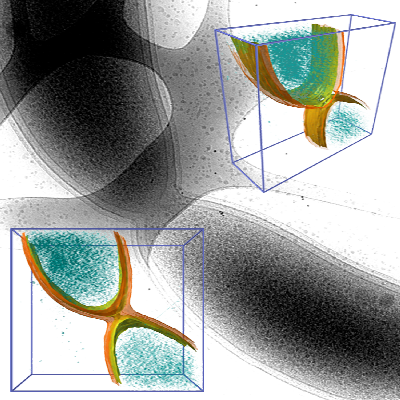
Bacteria contain a wealth of mechanisms that organize their internal and external structure. Electron Tomography offers the possibility to localize the cell components at a resolution that allows a better understanding of their structure and function. The 3D reconstruction of whole cells should lead to the ability to model spatial and temporal relationships among sub-cellular structures, organelles and macromolecular complexes.
In some cases the 3D reconstruction has a low signal to noise ratio resulting in data that produces difficult to distinguish visualization output. Volume data with features that are not easy to extract using automatic segmentation techinques make a perfect case for construction of a model of structural features. The model - discrete representation of structure partially visible in noisy data - is easier to visually interpret, making it easier for researchers to understand the shape and topology of such features.
In this work we show membrane models of dividing cells of Caulobacter Crescentus.
-
Web page for the self-guided demo
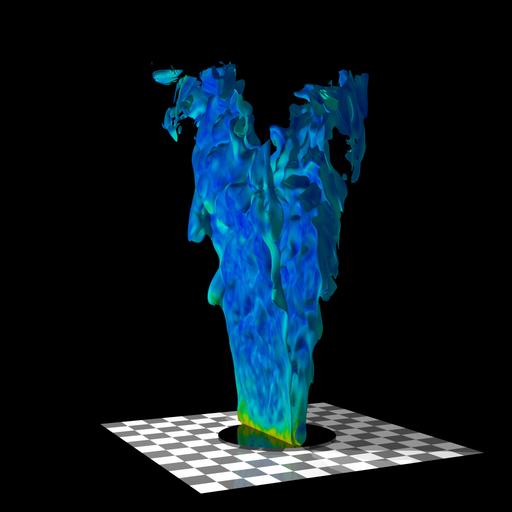
Combustion: Rod-stabilized V-flame
- Title:
Visualization of Rod-stabilized V-flames
- Description:
Visualization of a three-dimensional, time-dependent simulation of a laboratory-scale rod-stabilized premixed turbulent V-flame.
-
Web page for the self-guided demo.
DaVinci.nersc.gov
- Title:
Parallel Remote Visualization of AMR Combustion data using VisIt
- Description:
DaVinci is an SGI Altix 350 server running the SGI ProPack 4 64-bit Linux operating system, based on SUSE Linux Enterprise Server 9 (SLES9). Its main purpose is to provide visualization and data analysis capabilities to the NERSC user community. We will show the visualization of a 17GB dataset of an AMR three-dimensional simulation using DaVinci to run the parallel back-end of VisIt (LLNL) and a local workstation to run the client.
-
Web page for the self-guided demo.
Drosophilia Gene Expression Visualization
- Title: Drosophilia Gene Expression Visualization
- Description:
Together with the Berkeley Drosophila Transcription Network (BDTNP)
we have developed PointCloudXplore, a tool aimed at helping biologists
to understand the relationship between expression patterns of genes
in three dimensions. As part of this project, novel 3D point cloud
data sets are created from 3D confocal microscopy images containing
information about gene expression in fruit fly embryos at cellular
resolution. To support analysis of these high dimensional data sets,
PointCloudXplore integrates multiple views to ease analysis of complex
gene expression data. Each view emphasizes different data properties,
and interaction between the views makes it possible to perform detailed
analyses of the presented data. This type of interaction blends
high-dimensional information exploration with interactive,
3D visualization.
-
Web page for
self-guided demo.
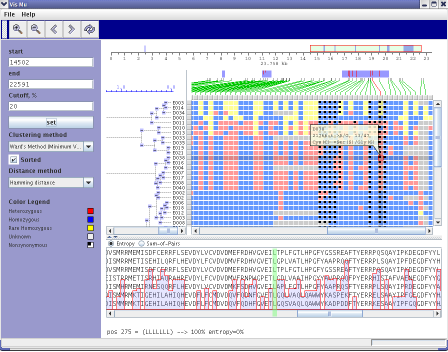
Interactive Single Nucleotide Polymorphism Visualization
|
- Title: Interactive Single Nucleotide Polymorphism Visualization
- Abstract:
Recent advances in sequencing technologies promise better diagnostics for
many diseases as well as better understanding of evolution of microbial
populations. Single Nucleotide Polymorphisms (SNPs) are established
genetic markers that aid in the identification of loci affecting
quantitative traits and/or disease in a wide variety of eukaryotic
species. With today's technological capabilities, it is possible to
re-sequence a large set of appropriate candidate genes in individuals with a given disease and
then screen for causative mutations. In addition, SNPs have been used
extensively in efforts to study the evolution of microbial populations,
and the recent application of random shotgun sequencing to environmental
samples makes possible more extensive SNP analysis of co-occurring and
co-evolving microbial populations. The program is available at
http://genome.lbl.gov/vista/snpvista.
- Web page for self-guided demo.
HDF5-FastQuery:
Accelerating Complex Queries on HDF Datasets using Fast Bitmap Indices
- Title: Accelerated Queries of HDF5 Datasets using Fast Bitmap Indices
- Abstract:
Efficient analysis of large scientific datasets often requires a means to
rapidly search and select interesting portions of data
based on ad-hoc search criteria. We present our work on integrating an
efficient searching technology named FastBit [2, 3]
with HDF5. The integrated system named HDF5-FastQuery allows the users to
efficiently generate complex selections on
HDF5 datasets using compound range queries of the form
(temperature > 1000) AND (70 < pressure < 90).
FastBit technology generates compressed bitmap indices that accelerate searches
on HDF5 datasets and can be stored together with
those datasets in an HDF5 file. Compared with other indexing schemes,
compressed bitmap indices are compact and very well
suited for searching over multidimensional data -- even for arbitrarily
complex combinations of range conditions.
- Web page for self-guided demo.
Talks
SC05 Talks