This is an archival copy of the Visualization Group's web page 1998 to 2017. For current information, please vist our group's new web page.
NOTE: HDF5-FastQuery is not yet available as a public release. We are working on improving the quality of the code so that we can contribute it to the HDF5 codebase.
Table of Contents
Introduction
Efficient analysis of large scientific datasets often requires a means
to rapidly search and select interesting portions of data
based on ad-hoc search criteria. We present our work on integrating an
efficient searching technology named FastBit [2, 3]
with HDF5. The integrated system named HDF5-FastQuery allows the users
to efficiently generate complex selections on
HDF5 datasets using compound range queries of the form
(temperature > 1000) AND (70 < pressure <
90). The FastBit
technology generates compressed bitmap indices that accelerate
searches on HDF5 datasets and can be stored together with
those datasets in an HDF5 file. Compared with other indexing schemes,
compressed bitmap indices are compact and very well
suited for searching over multidimensional data – even for arbitrarily
complex combinations of range conditions.
FastBit Indexing Technology
Bitmap indexing is a technique for processing complex, multi-dimensional ad-hoc queries on read-only data. They have been introduced into several commercial database systems by vendors such as Sybase, IBM and Oracle. FastBit is a specialized bitmap indexing technology for numeric data that uses a bitmap compression method designed to be more compute-efficient than the best available commercial implementations. The size of the compressed indices is typically about a third of the data size. FastBit facilitates very efficient multi-dimensional searches of scientific datasets.
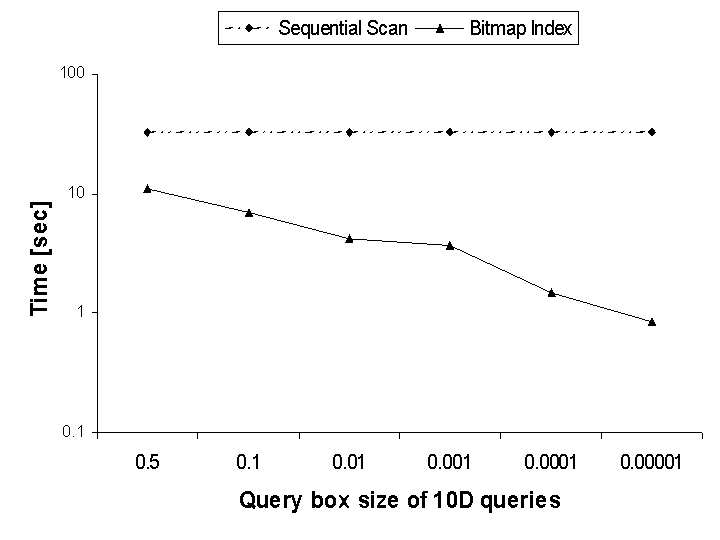
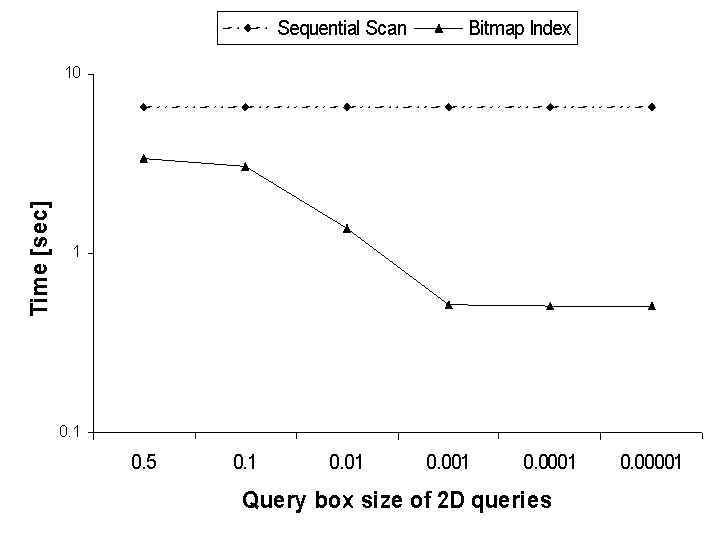
Figure 1 compares the performance of sequential scans to that of bitmap indices for processing multi-dimensional range queries. The sequential scan, where every element in the dataset must be evaluated against the query expression, is the most common approach for answering these types of queries in lieu of indexing technology. We see that the bitmap index significantly outperforms the sequential scan by a factor of 5 to 100, depending on the result size. We can also see that the smaller the query box size (i.e. the more selective the query), the higher is the performance advantage of the bitmap index.
 |
 |
HDF5-FastQuery
HDF5 supports complex selections based on multidimensional data coordinates (eg. hyperslab selection). HDF5-FastQuery extends the HDF5 selection mechanism to allow arbitrary range conditions on the data values contained in the datasets using the bitmap indices to accelerate the query. The FastQuery technology can efficiently support compound queries that span multiple datasets.
Our initial implementation uses a wrapper API that is designed to facilitate storage of time-series of multi-variable block- structured datasets which are common in the sciences. In the future, the storage organization can be expanded to accommodate more complex data schemas such as unstructured meshes, chemistry, and particle datasets. The API also allows us to seamlessly integrate the FastBit query mechanism for data selection with HDF5's standard hyperslab selection mechanism. Using the FastQuery API, one can efficiently select subsets of data from a HDF5 file using text-string queries.
 |
 |
 |
 |
The bitmap indices are stored in the same file as the datasets
they refer to and are opaque to the general HDF5 functions. A
query is posed to the API as a text-string such as "(temperature &ge
1000) AND (70 < pressure < 90)", where the names
specified in the range query correspond to the names of datasets in
the HDF5 file. The FastQuery interface will then consult
the stored bitmap indices that correspond to the specified dataset in
order to accelerate the selection of elements in the datasets
that meet the search criteria. An accelerated query on the
contents of a dataset requires only small portions of the
compressed
bitmap indices be read into memory, so extremely large datasets can be
searched with little memory overhead. The query engine
then generates an HDF5 selection that can be used to only read the
elements from the dataset that are specified by the query
string.
The FastBit technology is amenable to handling datasets and selections
that are far larger than system memory. In recent
experiments with data stored in 241Gigabyte data file[4], a search
that consumed 2467 seconds using sequential scan was
reduced to only 22.8 seconds using the bitmap indices. This same
ability to handle out-of-core data selections will be available
in the HDF5-FastQuery implementation.
Applications
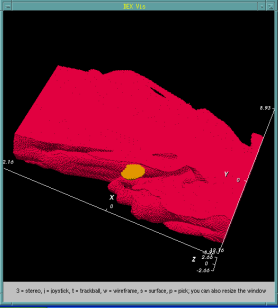
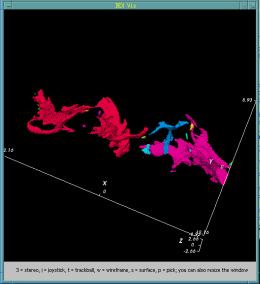
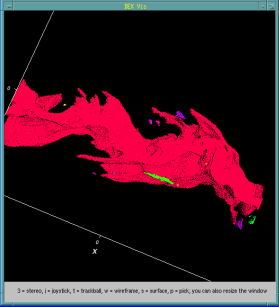
In addition to data analysis applications, we have applied bitmap indices for efficient query-based visualization within the DEX framework [1]. Figure 2 shows an interactive visualization process based on a large combustion simulation dataset. The example demonstrates how data is progressively interrogated to focus on cells that contain properties of interest.
References
- Kurt Stockinger, John Shalf, Wes Bethel, and Kesheng Wu. Query -Driven Visualization of Large Data Sets. In IEEE Visualization 2005, Minneapolis, MN, October 23-25, 2005, IEEE Computer Society Press.
- Kesheng Wu, Ekow J. Otoo, and Arie Shoshani. An Efficient Compression Scheme for Bitmap Indices. Technical Report LBNL-49626, Lawrence Berkeley National Laboratory, Berkeley, CA, 2002, To appear in ACM Transcations on Database Systems, 2006. ACM Press.
- Kesheng Wu, Ekow J. Otoo, and Arie Shoshani. On the Performance of Bitmap Indices for High Cardinality Attributes. In International Conference on Very Large Data Bases (VLDB), Toronto, Canada, August 31 - September 3, 2004, Morgan Kaufmann.
- K. Stockinger, K. Wu, S. Campbell, S. Lau, M. Fisk, E. Gavrilov, A. Kent, C.E. Davis, R. Olinger, R. Young, J.E. Prewett, P. Weber, T.P. Caudell, E.W. Bethel, S. Smith. ”Network Traffic Analysis With Query Driven Visualization SC 2005 HPC Analytics Results.” Accepted SC2005 HPC Analytics Challenge, August 2005.