Software

HDMF
Description: The Hierarchial Data Modeling Framework (HDMF) supports specification, creation, read, and write of scientific data formats. Originally developed as part of the NWB project, HDMF defines a reusable library for modeling scientific data and creation of data standardsAvailable at: https://hdmf-dev.github.io
Documentation: https://hdmf.readthedocs.io
Related Tools: 1) HDMF Common Schema, 2) HDMF Documentation Utilities, 3) HDMF Specification Language
Status: Active
Role: Project Lead and Developer
Citation: A. J. Tritt, O. Rübel, B. Dichter, R. Ly, D. Kang, E. F. Chang, L. M. Frank, K. E. Bouchard, "HDMF: Hierarchical Data Modeling Framework for Modern Science Data Standards," 2019 IEEE International Conference on Big Data (Big Data), Los Angeles, CA, USA, pp. 165-179, December, 2019, doi: 10.1109/BigData47090.2019.9005648.

PyNWB
Description: PyNWB is a Python package for working with NWB:N files. It provides a high-level API for efficiently working with Neurodata stored in the NWB:N format.Available at: https://github.com/NeurodataWithoutBorders/pynwb
Documentation: https://neurodatawithoutborders.github.io/pynwb
Related Tools: 1) NWB Schema, 2) MatNWB
Status: Active
Role: Project Lead and Developer
Citation: O. Rübel, A. Tritt, B. Dichter, T. Braun, N. Cain, N. Clack, T. J. Davidson, M. Dougherty, J.-C. Fillion-Robin, N. Graddis, M. Grauer, J. T. Kiggins, L. Niu, D. Ozturk, W. Schroeder, I. Soltesz, F. T. Sommer, K. Svoboda, L. Ng, L. M. Frank, and K. Bouchard. "NWB:N 2.0: An Accessible Data Standard for Neurophysiology," bioRxiv, January 17, 2019, doi: https://doi.org/10.1101/523035 (preprint)

Neurodata Extension Catalog
Description: The Neurodata Extensions Catalog (NDX Catalog) is a community-led catalog of extensions to the Neurodata Without Borders (NWB) data standar.Available at: https://nwb-extensions.github.io
Related Tools: 1) NDX Template, 2) Staged Extensions
Status: Active
Role: Project Lead
Parallel Peak Pruning
Description: Parallel Peak Pruning is the first shared SMP algorithm for fully parallel contour tree computation. PPP is available as part of the VTK-m library and supports parallel execution on CPUs and GPUs.Available at: https://gitlab.kitware.com/vtk/vtk-m
Status: Active
Role: Developer
Citation: H.Carr, G.H.Weber, C.Sewell, O. Rübel, P.Fasel and J.Ahrens, “Scalable Contour Tree Computation by Data Parallel Peak Pruning,” in IEEE Transactions on Visualization and Computer Graphics, Nov. 1, 2019, doi: 10.1109/TVCG.2019.2948616
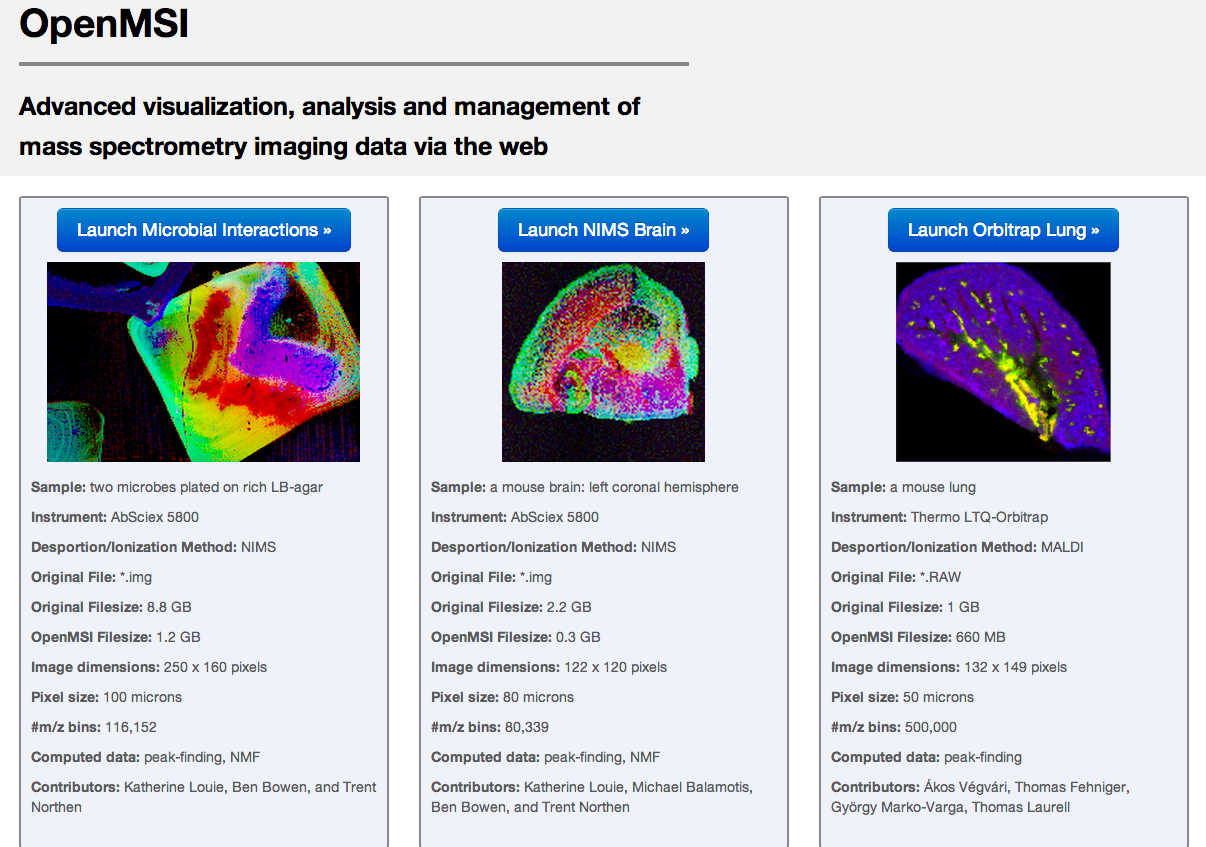
OpenMSI
Description: OpenMSI is an R&D100 award-winning, advanced application for web-based visualization, analysis, and management of mass spectrometry imaging data.Available at: www.openmsi.nersc.gov
Status: Active
Role: Compute Lead and Chief Software Architect
Patent applied for: Application number US 14/091,986
Licences ImaBiotech announced a license agreement with Lawrence Berkeley National Laboratory to “OpenMSI” intellectual property in April 2016 (see here)
Citation: Oliver Rübel, Annette Greiner, Shreyas Cholia, Katherine Louie, E. Wes Bethel, Trent R. Northen, and Benjamin P. Bowen, "OpenMSI: A High-Performance Web-Based Platform for Mass Spectrometry Imaging" Analytical Chemistry 2013 85 (21), 10354-10361, DOI: 10.1021/ac402540a. [BibTeX][Online at ACS]

BASTet
Description: BASTet is a novel framework for shareable and reproducible data analysis that supports standardized data and analysis interfaces, integrated data storage, data provenance, workflow management, and a broad set of integrated tools. BASTet has been motivated by the critical need to enable MSI researchers to share, reuse, reproduce, validate, interpret, and apply common and new analysis methods.Available at: https://openmsi.nersc.gov/openmsi/client/bastet.html
Status: Active
Role: Project Lead
First released: March 2016
Citation In review

WarpIV
Description: WarpIV is a python application that enables efficient, parallel visualization and analysis of simulation data while it is being generated by the Warp simulation framework. WarpIV integrates state-of-the-art in situ visualization and analysis using VisIt with Warp, supports management and control of complex in situ visualization and analysis workflows, and implements integrated analytics to facilitate query and feature-based data analytics and efficient large-scale data analysis.Available at: https://bitbucket.org/berkeleylab/warpiv
Status: Active
Role: Project Lead
Citation O. Rübel, B. Loring, J.-L. Vay, D. P. Grote, R. Lehe, S. Bulanov, H. Vincenti, and E.~W. Bethel, "WarpIV: In Situ Visualization and Analysis of Ion Accelerator Simulations," IEEE Computer Graphics & Applications, Scientific Visualization, pp. 22-35, May/June, 2016. LBNL-1005718. [Online] Available: http://ieeexplore.ieee.org/xpls/abs_all.jsp?arnumber=7466742 Supplemental Material: http://s3.amazonaws.com/ieeecs.cdn.csdl.public/mags/cg/2016/03/extras/mcg2016030022s1.pdf (Alternative download of PDF: https://publications.lbl.gov/islandora/object/ir%3A1005718)
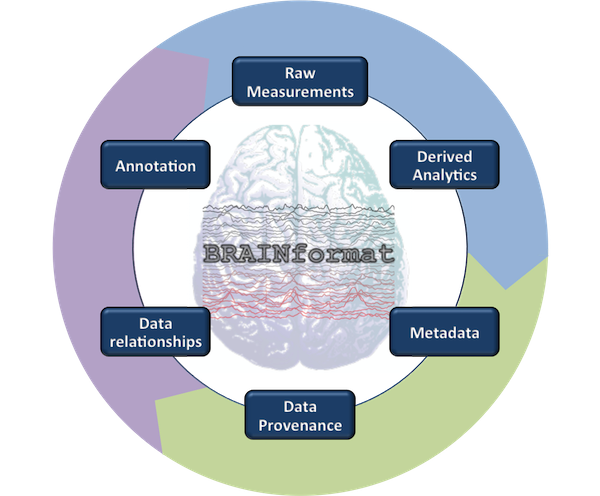
BRAINformat
Description: The LBNL BRAINFormat library specifies a general data format standardization framework and implements a novel file format for management and storage of neuro-science data. The library provides a number of core modules that can be used for implementation and specification of scientific application formats in general. Based on these components, the library implements the LBNL BRAIN file format.Available at: https://bitbucket.org/oruebel/brainformat
Status: Active
Role: Main Software Architect and Developer
Citation Oliver Rübel, Prabhat, Peter. Denes, David Conant, Edward Chang, and Kristofer Bouchard, "BRAINformat: A Data Standardization Framework for Neuroscience Data," in bioRxiv, Cold Spring Harbor Labs Journals, August 2015. DOI 10.1101/024521. Online
Metabolite Atlas
Description: Metabolite Atlas is a web-based atlas to liquid chromatography–mass spectrometry (LCMS) data.Available at: https://metatlas.nersc.gov/
Status: Active
Role: My main role in the project has been as a developer and adviser. The project is led by Benjamin P. Bowen (LBNL).
Contact: Benjamin P. Bowen, BPBowen@LBL.GOV
Citation Yushu Yao, Terence Sun, Tony Wang, Oliver Rübel, Trent Northen, and Benjamin P. Bowen, "Analysis of Metabolomics Datasets with High-Performance Computing and Metabolite Atlases," Metabolites 2015, 5, pp. 431-442, July 2015. DOI: 0.3390/metabo5030431
MAGI:
Description: MAGI is a tool to quickly find and score consensus between metabolite identifications and gene annotations.Available at: https://magi.nersc.gov/
Status: Active
Role: My main role in the project has been as a developer and adviser. The project is led by Benjamin P. Bowen (LBNL).
Contact: Benjamin P. Bowen, BPBowen@LBL.GOV
Citation O. Erbilgin, O. Rübel, K.B. Louie, M. Trinh, M. Raad, T. Wildish, D. Udwary, C. Hoover, S. Deutsch, T.R. Northen, B.P. Bowen. "MAGI: A Method for Metabolite Annotation and Gene Integration," ACS Chemical Biology, April 2019, 19;14(4):704-714. doi: 10.1021/acschembio.8b01107.
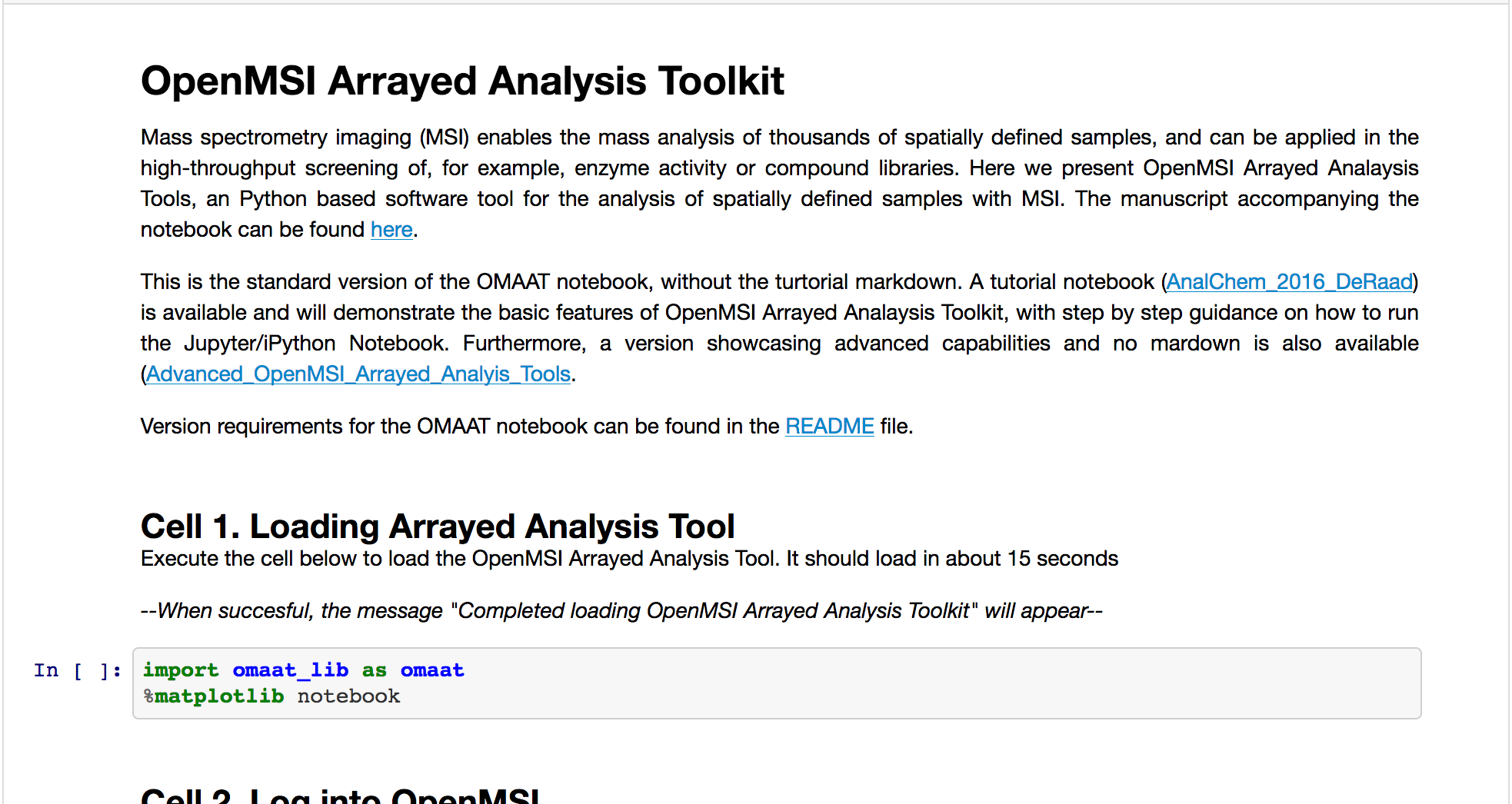
OMAAT: OpenMSI Arrayed Analysis Toolkit
Description: OpenMSI Arrayed Analysis Toolkit (OMAAT) is a new software package to analyze spatially defined samples in mass spectrometry imaging (MSI) using OpenMSI and Jupyter.Available at: https://github.com/biorack/omaat
Status: Active
Role: Developer
Contact: Benjamin P. Bowen, BPBowen@LBL.GOV
Citation (in review)
PointCloudXplore
Description: Developed at the Institute for Data Analysis and Visualization (IDAV), in collaboration with the BDTNP, PointCloudXplore is an advanced visualization tool for spatial and temporal 3D gene expression data. It was developed to help biologists understand the relationship between gene expression patterns in three dimensions. To support analysis of these high dimensional data sets, PointCloudXplore integrates multiple views to ease analysis of complex gene expression data. Each view emphasizes different data properties, and interaction between the views makes it possible to perform detailed analyses of the presented data. This type of interaction blends high-dimensional information exploration with interactive, 3D visualization.Overview: http://vis.lbl.gov/Vignettes/Drosophila/index.html
Available at: http://bdtnp.lbl.gov/Fly-Net/bioimaging.jsp?w=pcx
User manual http://bdtnp.lbl.gov/Fly-Net/pcx.jsp?w=vis
Status: PointCloudXplore was part of my Master and Ph.D. research. The project is currently inactive.
Role: Software Architect and Developer
Citation: Oliver Rübel, Gunther H. Weber, Soile V.E. Keränen, Charless C. Fowlkes, Cris Luengo Hendriks, Lisa Simirenko, N.Y. Shah, Michael B. Eisen, Mark D. Biggin, Hans Hagen, J. Damir Sudar, Jitendra Malik, David W. Knowles, and Bernd Hamann, "PointCloudXplore: Visual analysis of 3D gene expression data using physical views and parallel coordinates", in: Sousa Santos, B., Ertl, T. and Joy, K.I., eds., Data Visualization 2006 (Proceedings of EuroVis 2006), Eurographics Association, Aire-la-Ville, Switzerland, pp. 203-210
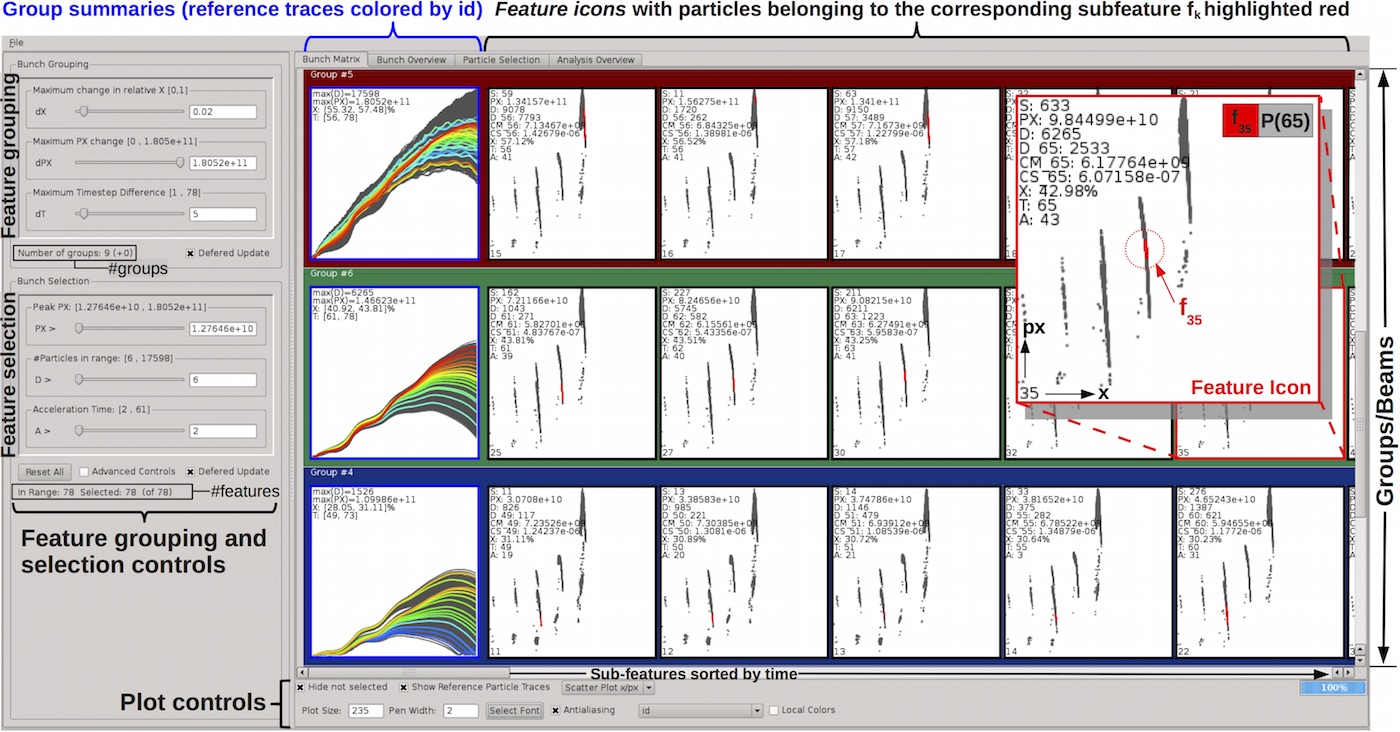
Feature-based Analysis of Plasma-based Particle Acceleration Data
Description: Plasma-based particle accelerators can produce and sustain thousands of times stronger acceleration fields than conventional particle accelerators, providing a potential solution to the problem of the growing size and cost of conventional particle accelerators. To facilitate scientific knowledge discovery from the ever growing collections of accelerator simulation data, we describe a novel approach for automatic detection and classification of particle beams and beam substructures due to temporal differences in the acceleration process. The automatic feature detection in combination with a novel visualization tool for fast, intuitive, query-based exploration of acceleration features enables an effective top-down data exploration process, starting from a high-level, feature-based view down to the level of individual particles.Available at: https://code.lbl.gov/projects/lwfapath/
Status: Inactive
Role: Main Software & Algorithms Architect and Developer
Citation: Oliver Rübel, Cameron G.R. Geddes, Min Chen, Estelle Cormier-Michel and E. Wes Bethel, "Feature-based Analysis of Plasma-based Particle Acceleration Data," IEEE Transactions on Visualization and Computer Graphics, 20(2):196–210, February 2014.
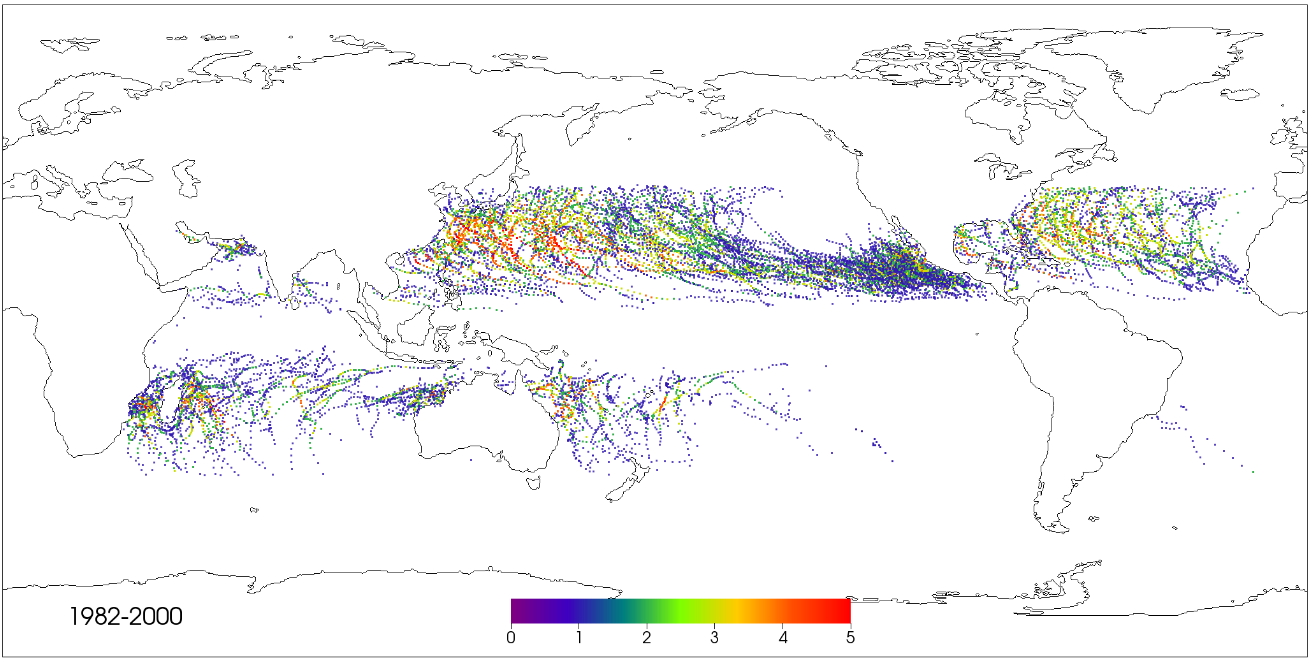
TECA: Toolkit for Extreme Climate Analysis
Description: TECA is a parallel toolkit for detecting extreme events in large climate database.Available at: Public release coming soon.
Status: Active. The development of TECA is currently being led by my colleague Burlen Loring.
Role: I was one of the main developers of the first version of TECA. I currently no longer play a major role in the development of TECA.
Citation: Prabhat, Oliver Rübel, Surendra Byna, Kesheng Wu, Fuyu Li, Michael Wehner and E. Wes Bethel, "TECA: A Parallel Toolkit for Extreme Climate Analysis," Third Worskhop on Data Mining in Earth System Science (DMESS 2012) at the International Conference on Computational Science (ICCS 2012), Omaha, Nebraska, June, 2012. LBNL-5352E (BibTeX)(PDF)
msr: Morse-Smale Approximation, Regression and Visualization
Description: Discrete Morse-Smale complex approximation based on kNN graph. The Morse-Smale complex provides a decomposition of the domain. This package provides methods to compute a hierarchical sequence of Morse-Smale complicies and tools that exploit this domain decomposition for regression and visualization of scalar functions.Available at: https://cran.r-project.org/web/packages/msr/index.html
Status: The primary developers of this software are Samual Gerber and Kristin Potter. This software was originally developed as part of a collaboration during my PostDoc at LLNL and is being maintained by LLNL and SCI.
Role: I contributed to the development of the original version of msr. I currently no longer play an active role in the development of msr.
Citation: Samual Gerber, Oliver Rübel, Peer-Timo Bremer, Valerio Pascucci and Robert T. Whitaker. "Morse-Smale Regression." Journal of Computational and Graphical Statistics (in press) (published online Jan.2012), LBNL-5682E, (BibTeX)(PDF)(article at JCGS)(MSR package for R available at CRAN here).

Other Software
- VisIt: VisIt is an open Source, interactive, scalable, visualization, animation and analysis tool. Over the course of my research I have contributed to the development of various components of VisIt, in particular the the design of Cumulative Selections, the integration of H5Part and FastBit, and features for particle tracing.
- AYLA Ayla is a free, open source visualization tool for researchers in biochemistry, molecular dynamics, and protein folding. Ayla has been primarily developed by William Harvey.. As part of my research at Lawrence Livermore National Laboratory I contributed to the design of some of the methods and I am a co-author on the corresponding paper: W. Harvey, I.-H. Park, O. Rübel, V. Pascucci, P.-T. Bremer, C. Li, and Y. Wang, “A Collaborative Visual Analytics Suite for Protein Folding Research,” Journal of Molecular Graphics and Modeling (JMGM), Vol. 53, pp.59–71, September 2014. [Online]. Available: http://www.sciencedirect.com/science/article/pii/S1093326314000990
- CASCADE: As part of my work with the exciting CASCADE project on "Calibrated And Systematic Characterization, Attribution, and Detection of Extremes" I worked on the development of API's for the design and implementation of climate workflows and data transfer.
- Particle Beam Analysis Over the course of my Ph.D. research I contributed to and led the development of several different approaches for the detection and analysis of particle beams in laser plasma accelerator simulations. (1) Daniela Ushizima, Cameron Geddes, Estelle Cormier-Michel, E. Wes Bethel, Janet Jacobsen, Prabhat, Oliver Rübel, Gunther Weber, Bernard Hamann, Peter Messmer, Hans Hagen. "Automated Detection and Analysis of Particle Beams in Laser-plasma Accelerator Simulations", In-Tech, 367-389, 2010. LBNL-3845E. (2) O. Rübel, C.G.R. Geddes, E. Cormier-Michel, K. Wu, Prabhat, G.H. Weber, D.M. Ushizima, P. Messmer, H. Hagen, B. Hamann, and E.W. Bethel, "Automatic Beam Path Analysis of laser Wakefield Particle Acceleration Data", IOP Computational Science & Discovery, 2 015005 (38pp), Nov, 2009, LBNL-2734E. and (3) see Feature-based Analysis of Plasma-based Particle Acceleration Data
- High-throughput Characterization of Porous MaterialsJihan Kim, Richard Martin, Oliver Rübel, Maciej; Haranczyk and Berend Smit, "High-throughput Characterization of Porous Materials Using Graphics Processing Units," Journal of Chemical Theory and Computation, 2012, 8 (5), pp 1684–1693, DOI: 10.1021/ct200787v, March, 2012, LBNL-5409E
- HPC for Finance Data Analytics E. Wes Bethel, David Leinweber, Oliver Rübel, Kesheng Wu (authors in alphabetical order), Federal Market Information Technology in the Post Flash Crash Era: Roles of Supercomputing, Workshop on High Performance Computational Finance at SuperComputing 2011 (SC11), LBNL-5263E
Data

Mass Spectrometry Imaging Data
Description Mass spectrometry imaging (MSI) is widely applied to image complex samples for applications spanning health, microbial ecology, and high throughput screening of high-density arrays. MSI has emerged as a technique suited to resolving metabolism within complex cellular systems; where understanding the spatial variation of metabolism is vital for making a transformative impact on science. Via OpenMSI, we and our users have made numerous MSI datasets available to the public. Available at: www.openmsi.nersc.gov